Drosophila Genomics Resource Center
Grant Number: P40OD010949
Research Emphasis/Objectives
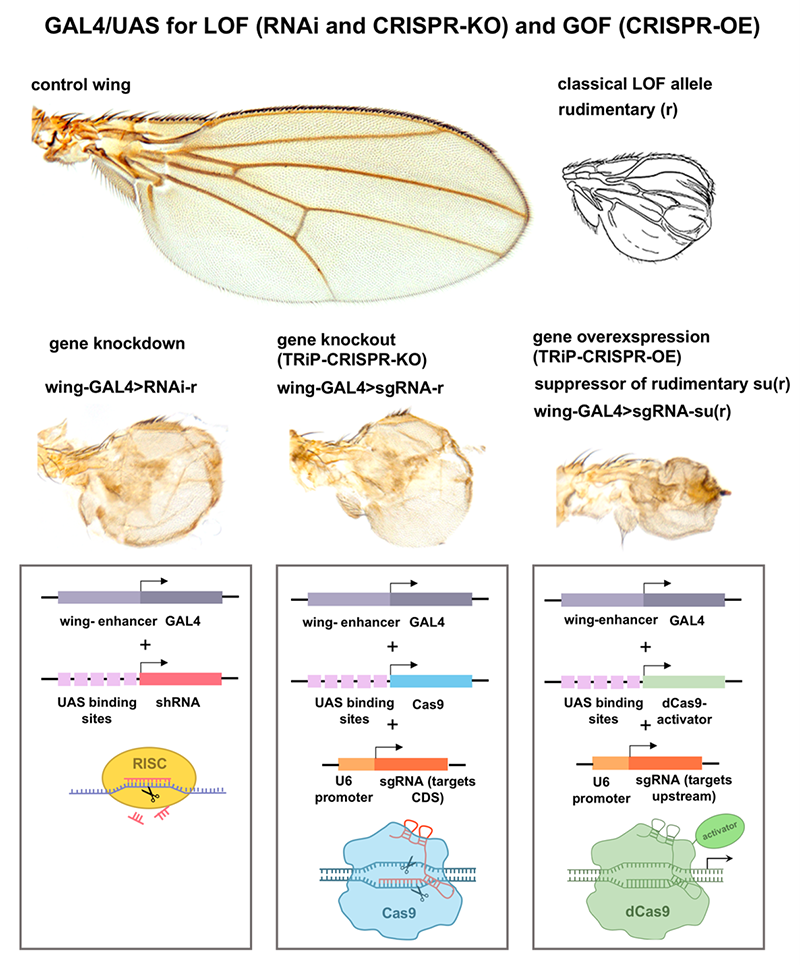
The Drosophila Genomics Resource Center (DGRC) supports an international community of scientists utilizing Drosophila melanogaster for biomedical research. The mission of the DGRC is to (1) provide the research community broad access to genomics resources by acquiring, archiving, curating, and distributing genomics resources, including, clones, vectors, and cell lines; (2) facilitate the effective use of the genomics resources by providing guidelines and support; and (3) improve the genomics resources and protocols available to the research community. By balancing the process of archiving and curating material, efficient distribution, and economical access, the effort of the DGRC increases both the scientific rigor and reproducibility of the collective work of the research community and protects against the loss of vital materials.
Services Provided
- Collecting and distributing cDNA clones and vectors
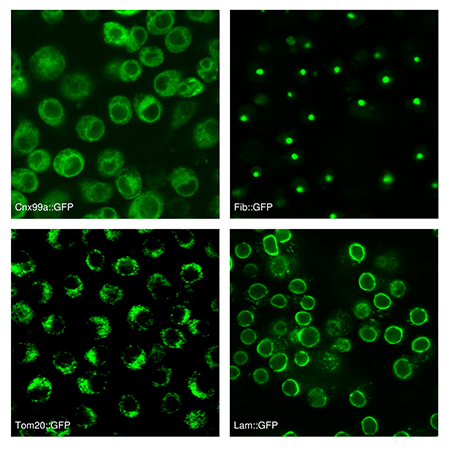
- Collecting and distributing Drosophila cell lines
- Developing and improving cell line/genomics technologies for use in Drosophila
- Assisting the research community in the use of these resources
Contact Information
Drosophila Genomics Resource Center (DGRC)
1001 E. Third St.
Bloomington, IN 47405
dgrc.bio.indiana.edu/Home
Principal Investigators
Andrew C. Zelhof, Ph.D.
Phone: 812-855-0294
azelhof@indiana.edu
Co-Principal Investigators
Kris Klueg, Ph.D.
Phone: 812-855-5510
dgrc@indiana.edu
Arthur Luhur, Ph.D.
Phone: 812-855-5510
dgrc@indiana.edu
Daniel Mariyappa
Phone: 812-855-5510
dgrc@indiana.edu
- Read more about Drosophila Genomics Resource Center
Log in to post comments




 Research Emphasis/Objectives
Research Emphasis/Objectives