Next-generation Drosophila Cell Lines to Elucidate the Cellular Basis of Human Diseases
Next-generation Drosophila Cell Lines to Elucidate the Cellular Basis of Human Diseases
Grant Number: R24OD019847
 Research Emphasis/Objectives
Research Emphasis/Objectives
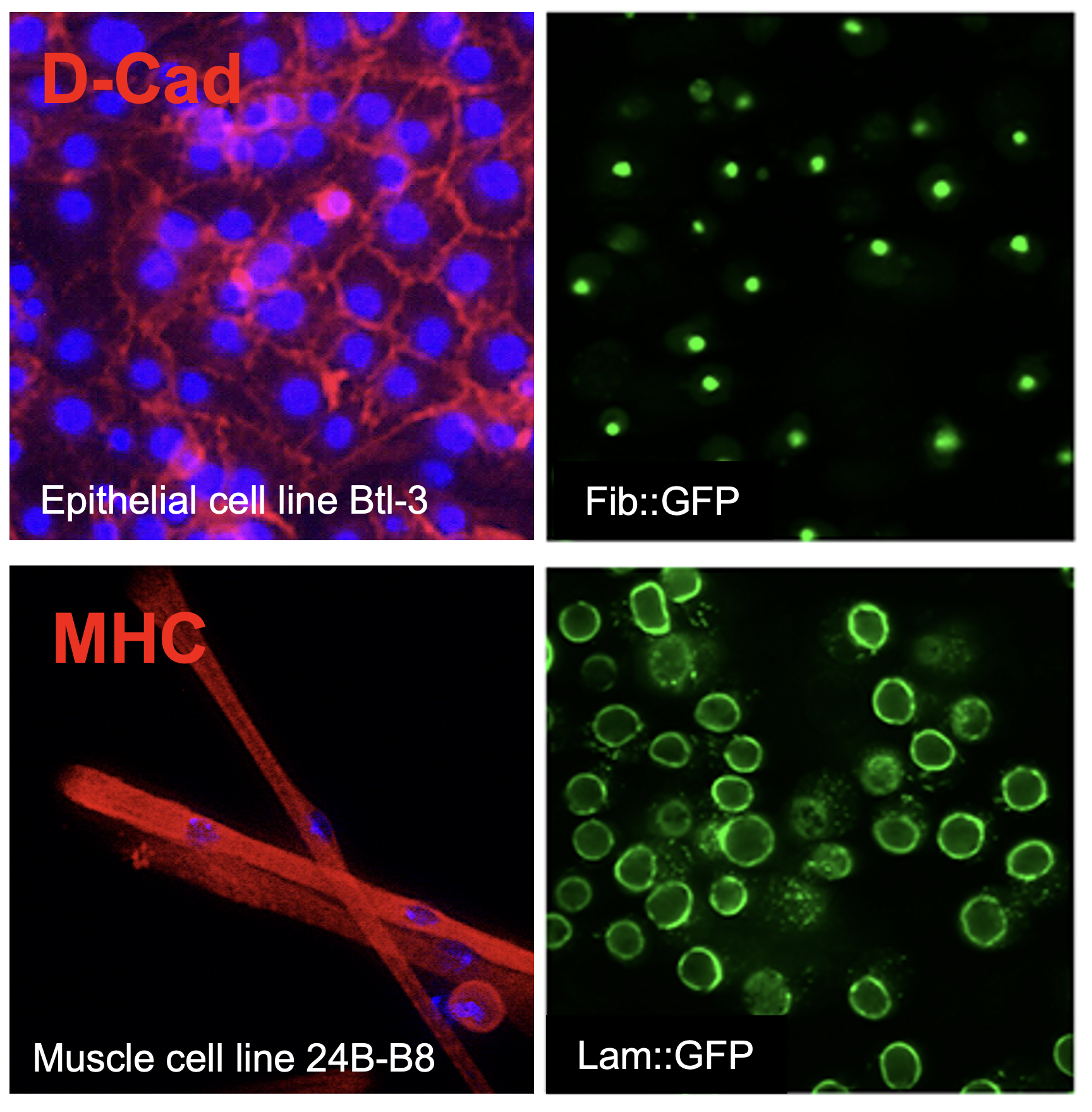
All diseases result from disturbances at the cellular level, and functional analyses of human diseases increasingly rely on cell biological studies to uncover root causes of disease. Cell-level analyses are revealing that many diseases affect the function and/or morphology of organelles, signal transduction, homeostasis, or cell growth. Resources that enable biological analyses in various cell types and the use of various assays are critical to assess the functions of genes implicated in diseases. Analysis of the Drosophila genome sequence has revealed striking conservation among fly and human genes. Drosophila orthologs have been identified for about two-thirds of human disease genes, and all major signal transduction pathways are conserved between flies and humans. The full potential of Drosophila cell lines, particularly for disease-related studies, has not yet been realized because the repertoire of cell lines has been limited, as is the availability of live-cell reporters and mutant cell lines. To meet this need, the resource used state-of-the-art technologies to generate reagents that will enable the community to fully exploit the power of Drosophila cell lines to address disease-related questions. Specifically, the resource (1) generated new continuous cell lines from specific Drosophila cell types using a technique pioneered by Dr. Amanda Simcox and characterized new cell lines with key attributes of neuronal, glial, muscle, blood, and epithelial cells; (2) used CRISPR-Cas9 genome engineering to develop a resource of cell lines expressing fluorescent markers of specific subcellular components, such as the nucleus or mitochondria; and (3) used CRISPR-Cas9 gene editing to generate knockout (loss-of-function) cell lines for a set of tumor suppressor genes and other genes. The cell lines should be useful for small-scale and large-scale studies, including genome-wide screens.
Services Provided
The cell line resources generated in this project were shared with the Drosophila Genomics Resource Center (DGRC) for distribution to the community.
Contact Information
Stephanie Mohr, Ph.D.
Director of Drosophila RNAi Screening Center/TRiP Functional Genomics Resources
Harvard Medical School
New Research Building, Room 336
77 Avenue Louis Pasteur
Boston, MA 02115
[email protected]
Resource name: Drosophila RNAi Screening Center (DRSC) modified cell line collection at DRSC/TRiP Functional Genomics Resources
Website (CRISPR-modified cell line information at the DRSC): fgr.hms.harvard.edu/crispr-modified-cell-lines
Website (new and CRISPR-modified cell line distribution by DGRC): dgrc.bio.indiana.edu/cells/Catalog
Co-Principal Investigators
Norbert Perrimon, Ph.D.
Harvard Medical School
Phone: 617-432-7672
[email protected]
Amanda Simcox, Ph.D.
Ohio State University
Phone: 614-292-8857
[email protected]
Other/Resource Contacts
Stephanie Mohr, Ph.D.
Co-Investigator and Director of DRSC/TRiP Functional Genomics
[email protected]



