A Resource for Developmental Regulatory Genomics
Grant Number: R24OD023046
Research Objectives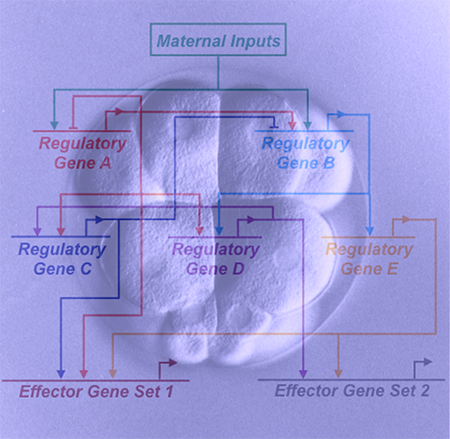
This award supports the creation and dissemination of research tools that will enable biologists to elucidate the genomic control of embryogenesis through the experimental dissection of developmental gene regulatory networks in sea urchins and other echinoderms.
Services Provided
(1) Production of recombineered bacterial artificial chromosomes (BACs) that encode fluorescent reporter proteins under the cis-regulatory control of endogenous echinoderm genes. (2) Digital analysis of gene expression by means of NanoString technology. (3) Resources for the genome-wide identification of cis-regulatory modules (CRMs). (4) Photoactivatable morpholino antisense oligo nucleotides for conditional (spatially and temporally regulated) gene knockdowns.
Contact Information
Resource for Developmental Regulatory Genomics
Dr. Charles Ettensohn
Department of Biological Sciences
Carnegie Mellon University
4400 Fifth Avenue
Pittsburgh, PA 15213
echinobase.org
Co-Principal Investigators
Dr. Charles Ettensohn
Phone: 412-268-5849
Fax: 412-268-7129
[email protected]
Dr. Veronica Hinman
Phone: 412-268-9348
Fax: 412-268-7129
[email protected]



