Supercomputing Technologies Funded by ORIP Helped Spur Approval of COVID-19 Vaccines and Save Lives*
Supercomputing has been gaining attention for its applications across biomedical research. A type of high-performance computing, this technology makes use of multiple central processing units (CPUs), thus allowing researchers to perform numerous calculations simultaneously for large-scale analysis. Researchers have used supercomputers to visualize a diverse range of biomedical data sets—from the intricate structures of a protein to complex genetic characteristics across global populations. ORIP is helping further progress in this area by funding shared resources for supercomputing through the S10 Instrumentation Grant Programs.
One such institution, the Fred Hutchinson Cancer Center (Fred Hutch), received funds from ORIP (S10OD028685) in 2020 to upgrade its high-performance computing cluster, which was established in 2004 and has been upgraded several times through the S10 programs and other mechanisms. The cluster provides researchers access to high-performance supercomputing resources that enhance their research, and the upgrades have increased the cluster’s data-processing capacity and overall performance.
The S10 funds were awarded to Fred Hutch in March 2020, just at the onset of the COVID-19 pandemic. A team of researchers from the Fred Hutch Vaccine and Infectious Disease Division used the supercomputing cluster to identify and validate immunological markers that have been used to predict the efficacy of four COVID-19 vaccines.1,2 They found that antibodies are consistently a correlate of protection in vaccinated individuals who were enrolled in Phase III trials of the Moderna, AstraZeneca, Janssen, and Novavax vaccines (Figure 1).
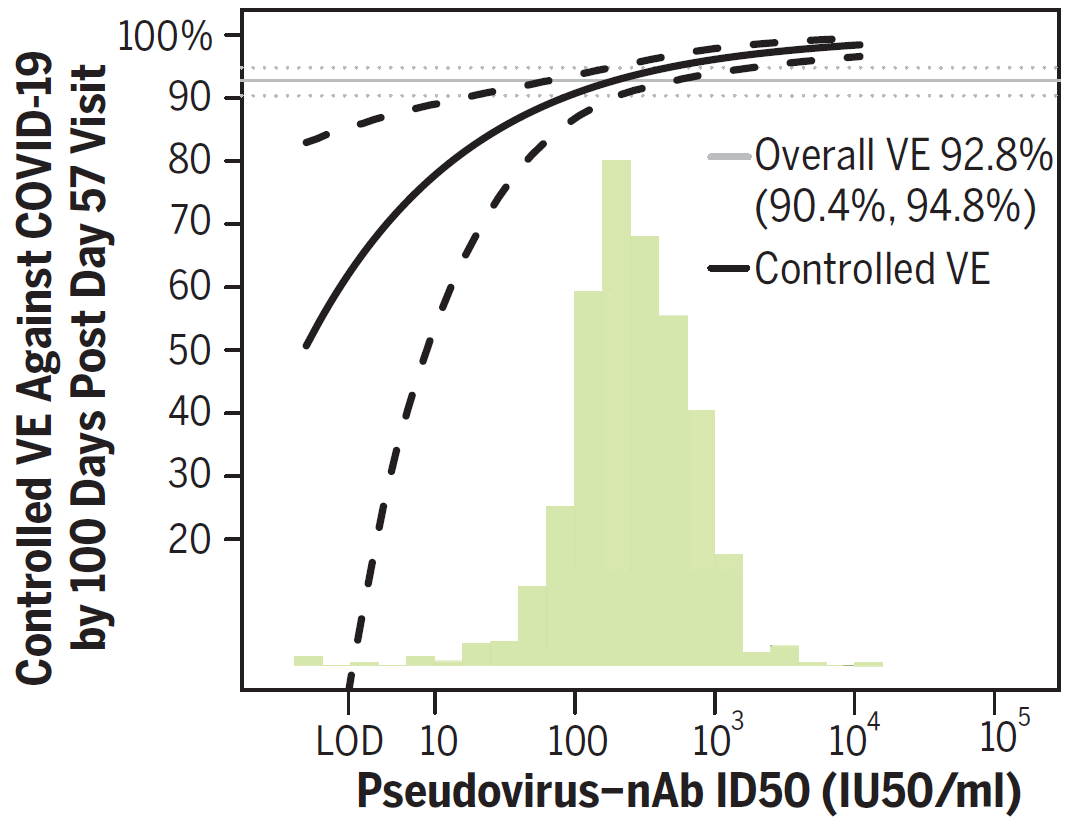
Dr. Peter Gilbert, a Professor in the Vaccine and Infectious Disease Division at Fred Hutch, explained that the S10-funded cluster helped inform timely decisions about vaccine approval and use. The research team characterized the correlates of protection using used statistical and machine-learning methods that were developed through Monte Carlo simulation studies, which Dr. Gilbert described as “computationally intensive.” Ultimately, the quick determination of vaccine efficacy was critical to saving lives during the COVID-19 pandemic.
Fred Hutch investigators perform research focused on eradicating cancer and other diseases. Their work involves computationally intensive statistical and machine-learning approaches that can be applied to studies of various topics, including cancer genomics, population-level risk prediction, vaccine development, microbiome analysis, protein structure prediction, and pharmacological design. Many Fred Hutch researchers pivoted quickly during the pandemic to perform studies on SARS-CoV-2, with a focus on viral evolution and vaccine development. They generated a large amount of data in a short time through these intensive efforts, and the upgraded cluster was essential for analyzing the results.

The upgraded cluster contains 7,000 CPU cores and 150 graphics processing units (Figure 2). The system operates using Slurm, an open-source workflow management software that uses a best-fit algorithm to optimize task assignments as they are submitted by users. Dr. Phil Bradley, a Professor in the Fred Hutch Public Health Sciences Division and Program Head of the Herbold Computational Biology Program, explained that nearly every Fred Hutch investigator has benefitted—either directly or indirectly—from the cluster resources.
Dr. Bradley emphasized that the cluster helps researchers analyze large data sets that are generated from these studies. “Data-intensive biological experiments are growing more and more, and large-scale computing helps researchers to get the most out of those experiments,” he explained. The established workflows and staff support within the cluster enable Fred Hutch researchers—even those without computational expertise—to access cutting-edge technologies for their studies. Additionally, by creating workflows that can be shared among research teams, investigators can leverage supercomputing technologies to improve the reproducibility of their research.
Looking forward, Dr. Bradley emphasized that supercomputing will emerge as an essential technology across the biomedical research enterprise. “I think the importance of cluster computing is only growing as our ability to generate data grows,” he reflected. “This is such a critical technology, and I think it’s really worthwhile supporting as a continued investment. We are building a community of researchers who are all knowledgeable about using this technology and who can help to educate new users.”
ORIP has helped support numerous advancements in supercomputing through the S10 programs. These funds have supported various areas of research—including image-based phenotyping, genome-wide association studies, evolution of pathogens, and large-scale studies of the brain—at institutions across the United States. ORIP’s S10 programs support purchases of commercially available instruments to enhance research by NIH-funded investigators. Each instrument funded through the S10 programs must be shared among multiple users, which ensures efficient use of instruments for research operations, enhances cost-effectiveness of NIH’s investment, and maximizes benefits for thousands of investigators in hundreds of institutions nationwide.
For more information on ORIP’s S10 Programs, please visit https://orip.nih.gov/division-construction-instruments/s10-instrumentation-programs.
[* This Research Highlight has been selected as a flip card published on the Impact of NIH Research page.]
References
1 Gilbert PB, Montefiori DC, McDermott AB, et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science 2022;375(6576):43–50. doi:10.1126/science.abm3425.
2 Gilbert PB, Donis RO, Koup RA, et al. A COVID-19 milestone attained—a correlate of protection for vaccines. The New England Journal of Medicine 2022;387(24):2203–2206.



