Solving the Mystery of Crystalline Protein Structures
Although X-ray crystallography was invented more than 100 years ago, it is still one of the most powerful techniques used to determine the three-dimensional (3D) structure of proteins. Technological advances during the last 10 years, such as the development of pixel array detectors (PADs), have improved the speed and quality of data collection, which in turn allows for the more accurate determination of 3D crystal structures. Knowing the 3D structure of a protein provides scientists with important information that can allow them to determine the function and biology of the molecule. For example, proteins that play a role in cellular function can be studied to determine whether they contribute to disease pathology. If so, then scientists can use knowledge of the structure to chemically identify agents that bind to the protein and to alter their action. Drugs then can be designed to treat various diseases, including cancer and autoimmune and infectious diseases.
Determining the structure of a protein is a complicated process that requires sophisticated equipment and knowledge of chemistry and biochemistry. First, scientists use their expertise to grow crystals in special conditions. Once the complicated process of creating a crystal has been completed, the researcher must verify its quality. Typically, onsite low-power X-ray scattering systems are used to screen crystals to ensure that the samples are of high quality and free of impurities. These systems also can be used to provide low-resolution readouts, which may answer some questions researchers have about the structure and related chemical properties. To obtain high-resolution structures, scientists need to use a synchrotron facility, where more advanced X-ray detectors, such as PADs, must be used for data collection.
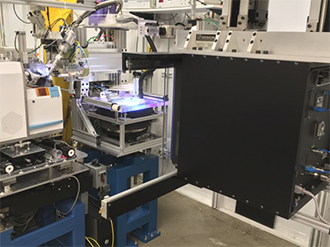
Both low-power X-ray scattering systems and high-intensity synchrotron X-rays bombard the prepared crystal with X-rays to generate raw data about the protein. In addition to high-quality samples, high-resolution crystallography also requires an intense, focused X-ray beam for bombardment and high-performance detectors to yield a clear diffraction pattern. Scientists use the resulting diffraction pattern, which shows the position of each atom in the protein, in the complicated visualization process. PADs, which digitally count the number of photons emanating from the protein, provide higher signal to noise, higher quantum efficiency detection (i.e., increased effectiveness), a much higher effective dynamic range, and a significantly faster data collection rate compared to previous detector technologies. Dr. Ealick, who has been using PAD technology for approximately 5 years, explains that the PAD readout time takes only milliseconds, which is approximately 1,000 times faster than older technology. The PAD’s low electronic noise allows a much clearer, higher resolution 3D structure than was previously attainable. The final 3D image ultimately is more than the sum of its data, allowing scientists to see the structure and its biochemistry, such as the motion of different helices or amino acid residues in the active site of the protein, to elucidate its biological function.
The National Institutes of Health’s (NIH) Office of Research Infrastructure Programs (ORIP) enables such research by supporting infrastructure that provides scientists with the resources required to answer specific research questions. In particular, ORIP has used its S10 Shared Instrumentation Grant program to support X-ray diffraction technology and has awarded instruments to several laboratories and facilities. Specifically, the NIH supported the installation of low-power X-ray scattering systems at university facilities and 10 PADs in synchrotron facilities across the United States. One example is an award to Dr. Steven Ealick from Cornell University of a PAD installed at the Advanced Photon Source at the Argonne National Laboratory.
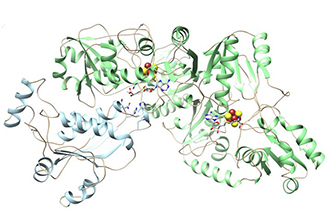
The PAD at Argonne annually serves approximately 70 NIH-supported projects and hundreds of investigators in essentially all areas of structural biology. In just the first few months of 2017, five peer-reviewed journal articles have reported structures whose determination would not be possible without the PAD awarded through NIH’s S10 program. For example, researchers reported the crystal structures of two polynucleotide ligases that form a protein complex. They repair DNA and RNA damage and therefore are essential in maintaining genomic integrity. These ligases are excellent targets for developing drugs for infectious diseases and cancer.1 Another group of researchers using the NIH-awarded PAD found that S-adenosyl-l-methionine (SAM) enzymes are a key component of the detoxification system to metabolize and clear xenobiotics—such as environmental toxins and pharmaceutical and complex food compounds—in humans. Understanding the structures of SAM enzymes will help researchers study the association between the detoxification system and xenobiotics-influenced diseases, such as cancer and neurological and immune diseases.2 The NIH-awarded PAD also was instrumental in determining the crystal structures of two important and conserved human enzymes involved in cancer mutagenesis and viral infection. Knowledge of these structures will allow the design of inhibitors to hinder viral and tumor evolvability.3 Finally, Dr. Ealick’s laboratory, in conjunction with a group at Yale University, recently used the PAD to determine the structure of viperin, an antiviral protein induced by the immune system molecule interferon, which has provided important clues to its function. The structure is in the process of being published.

Another important scientific discovery relied on two S10 grants, one for a low-power X-ray scattering system awarded to Columbia University and one for the PAD at Argonne. The recently reported breakthrough illustrates the tandem use of two NIH-supported instruments. The research team solved challenging high-resolution atomic structures of full-length yeast separase and its complex formation with the inhibitor securin. Separase is a large protein that plays a crucial role in chromosome segregation during cell division and its activity is regulated by securin. Because separase is overexpressed in cancer cells, determining the structure of the separase-securin complex could provide insight into its molecular mechanisms, eventually leading to the identification of potential drug targets to inhibit tumor cell proliferation.4 These are just a few examples of the exciting research made possible through NIH’s S10 Shared Instrumentation Grant program.
ORIP supports a broad community of researchers, providing them with sophisticated techniques and instruments that lead to important scientific discoveries. The research that leads to these discoveries is a long, complex process that requires a great deal of knowledge and expertise in addition to powerful equipment. ORIP is pleased that researchers recognize the benefits of the S10 Shared Instrumentation Grant program, whose investment makes research efforts possible.
References
1 Unciuleac MC, Y Goldgur, and S Shuman. Two-metal versus one-metal mechanisms of lysine adenylylation by ATP-dependent and NAD+-dependent polynucleotide ligases. Proc Natl Acad Sci U S A. 2017 Mar 7;114(10):2592–2597.
2 Bruender NA, TA Grell, DP Dowling, RM McCarty, CL Drennan, and V Bandarian. 7-carboxy-7-deazaguanine synthase: A radical S-adenosyl-L-methionine enzyme with polar tendencies. J Am Chem Soc. 2017 Jan 3;139(5):1912–1920.
3 Shi K, et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nature Structural & Molecular Biology. 2017 Feb;24(2):131–139.
4 Luo S and L Tong. Molecular mechanism for the regulation of yeast separase by securin. Nature. 2017 Feb 9;542(7640):255–259.



