S10 Instrumentation Programs Enhance Use of Cryo-EM Technologies for COVID-19 Research
Cryogenic electron microscopy (cryo-EM) can offer unique insights on biological molecules, enabling investigators to piece together complex structures. Two awards to the University of California, San Francisco (UCSF), through ORIP’s S10 Shared Instrumentation Programs, S10OD020054 and S10OD021741, have provided the infrastructure necessary for innovative research using this technology.
The awards funded the acquisition of a Talos Arctica 200kV cryogenic transmission electron microscope equipped with a Gatan direct electron detection camera, as well as a high-performance Linux computer cluster that is dedicated to processing the large data sets generated during single-particle cryo-EM experiments.
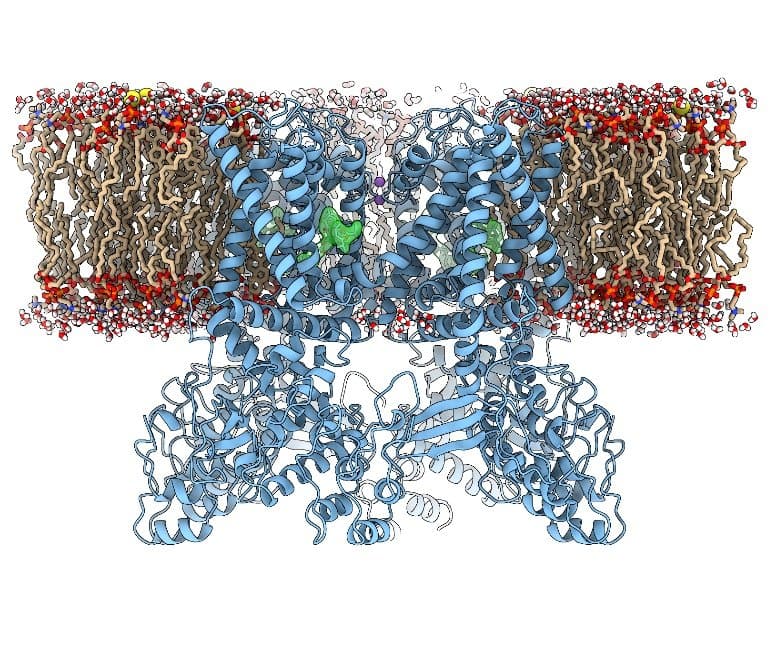
Dr. Yifan Cheng, Professor of Biochemistry and Biophysics at UCSF and Investigator at Howard Hughes Medical Institute, explained that technological breakthroughs in cryo-EM have led to unprecedented, rapid progress within the field of structural biology. His laboratory has used the technology to determine the structure of transient receptor potential cation channel subfamily V member 1 (TRPV1), a membrane channel protein (Figure 1), in collaboration with Dr. David Julius, Professor and Chair of Physiology at UCSF. This was the first atomic structure of a membrane protein to be determined by this method. The team reported their findings in two 2013 Nature articles1,2 that were highly cited in subsequent years.
“This was the first atomic structure of a membrane protein with unknown structure determined by single-particle cryo-EM,” Dr. Cheng explained. “Because [TRPV1] is a membrane protein, most thought [determining the structure] was impossible by single-particle cryo-EM. It was a big surprise to the field and promoted everyone to shift to cryo-EM.”
Traditionally, biologists have determined molecular structures using X-ray crystallography. In this technique, the protein is purified and set in hundreds to thousands of conditions within a crystallization tray to screen for the right condition that allows the growth of crystals. Once a well-ordered crystal is obtained, the crystal then is taken to a synchrotron beamline to collect diffraction data, which are used to determine the protein’s structure.
Dr. Cheng explained that X-ray crystallography is well established, but the process of crystallization imposes considerable drawbacks for investigators. Crystallization, he emphasized, is a process that could take a single day or decades to complete. Cryo-EM allows investigators to bypass the crystallization process.
With this new technique, the molecule is frozen in a single layer of amorphous ice by rapid cooling, and hundreds to thousands of images of frozen molecules are taken via the electron microscope. These images then are aligned computationally, allowing the generation of a high-resolution structure. These new capabilities have been crucial for numerous discoveries, including the determination of the spike protein of SARS-CoV-2, the virus that causes COVID-19.
“There are so many structural biology questions that, in the past, the available technology made them difficult to address,” Dr. Cheng emphasized. “Now suddenly, all of [these questions] can be a feasible target. Everyone is trying to get their hands on the new instrument and learn how to do these things. ... The whole field has completely shifted.”

The Arctica 200kV is among UCSF’s most productive S10-funded instruments and has led to 15–20 papers per year over the past several years. It serves an estimated 150–200 users, many of whom are graduate students and postdoctoral fellows (Figure 2). Dr. Cheng emphasized that the S10 Instrumentation Programs have provided many investigators—including the structural biology community at UCSF—with new capabilities to determine protein structures more quickly and at high resolution.
Dr. Cheng reflected that during the SARS outbreak in 2003, crystallization of the spike SARS-CoV-1 protein took years to complete. In 2020, however, the structure of the new spike protein was determined by using cryo-EM in only 5 weeks after the first SARS-CoV-2 genome was released. This new knowledge has been crucial for the rapid development of vaccines and therapeutics for COVID-19 during the pandemic.
Dr. Cheng explained that at the beginning of the COVID-19 pandemic, all other research facilities were closed to investigators at UCSF. The cryo-EM instrument, however, is automated, allowing remote collection of data. This design enabled users to maintain productivity during this time. For example, the structural biology community at UCSF organized the Quantitative Biosciences Institute Coronavirus Research Group, a consortium in which scientists—most of them graduate students and postdoctoral fellows—were grouped into teams focused on structural studies of different SARS-CoV-2 proteins. This concerted effort has led to several high-profile publications, including papers in Science3 and Cell.4
Dr. Cheng expressed appreciation to the NIH for its support of structural biology research. He emphasized the impact of cryo-EM to both the research community and the general public. “This time, I really feel like we did something—the structural biology community did something for the entire society in fighting the COVID-19 pandemic,” Dr. Cheng reflected. “We feel great that we can make these contributions.”
References
1 Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 2013;504(7478):107–112. doi:10.1038/nature12822.
2 Cao E, Liao M, Cheng Y, Julius D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 2013;504(7478):113–118. doi:10.1038/nature12823.
3 Gordon DE, Hiatt J, Bouhaddou M, et al. Comparative host–coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 2020;370(6521):eabe9403. doi:10.1126/science.abe9403.
4 Asarnow D, Wang B, Lee WH, et al. Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia. Cell 2021;184(12):3192–3204. doi:10.1016/j.cell.2021.04.033.



