Reproducibility of Rodent Models for Human Disease: MiniMUGA as a Critical Genetic Tool
Mice are a crucial resource for the scientific community. Scientific researchers from numerous disciplines use the mouse as a model to mimic and recapitulate diseases and test scientific questions related to human health and disease. These efforts include basic research, studies on the pathogenesis of disease, therapeutics to treat disease, markers for the diagnosis of disease, and strategies (e.g., vaccines) to prevent disease (Figure 1).
An increasing number of scientific articles report that the phenotype of a given single-gene mutation in mice is modulated by the genetic background of the inbred strain in which the mutation is maintained. This effect is attributable to so-called modifier genes, which act in combination with the causative gene.
Understanding background genetics in mice is crucial to characterize their responses to experimental treatments. This information also is needed for scientific reproducibility—the ability to replicate experiments across laboratories. Researchers typically properly document many underlying factors—such as health and environment—but often fail to consider the effect of genetics in reproducibility. A new iteration of the Mouse Universal Genotyping Array (i.e., MiniMUGA) was developed by researchers at The University of North Carolina at Chapel Hill (UNC) under a contract with NEOGEN Corporation to bridge this knowledge gap across the realm of research using mice, providing researchers key information on the genetic features of laboratory strains.
Traditionally, most laboratory mice are inbred, meaning that they are homozygous (i.e., possessing two identical alleles) at each gene locus. When working with inbred strains, researchers can more accurately predict the genetic outcomes of their specimens. Many researchers therefore assume that these genomes will remain identical indefinitely. Dr. Fernando Pardo Manuel de Villena, Professor and Chair in the Department of Genetics at the UNC School of Medicine, revealed that this is not the case.
Spontaneous mutations, as well as human errors, can lead to genetic changes in the colonies over time, Dr. Pardo Manuel de Villena explained. Without proper documentation, these factors can lead to misinterpretation of experimental findings. The results of such studies are simply anecdotes, findings that cannot be placed in the larger context of biomedical research.
“Anecdotal findings are interesting for the lay press but are not enough for science,” Dr. Pardo Manuel de Villena stated. “That is lazy thinking and bad use of limited resources. If you have not thought about these things, you are not doing really good science.”
Enhancing Rigor, Transparency, and Translatability in Animal Research

The challenge of maintaining rigor and reproducibility is relevant to all areas of NIH-funded animal research. In June 2021, the NIH Advisory Committee to the Director Working Group on Enhancing Rigor, Transparency, and Translatability in Animal Research published a report addressing this issue.
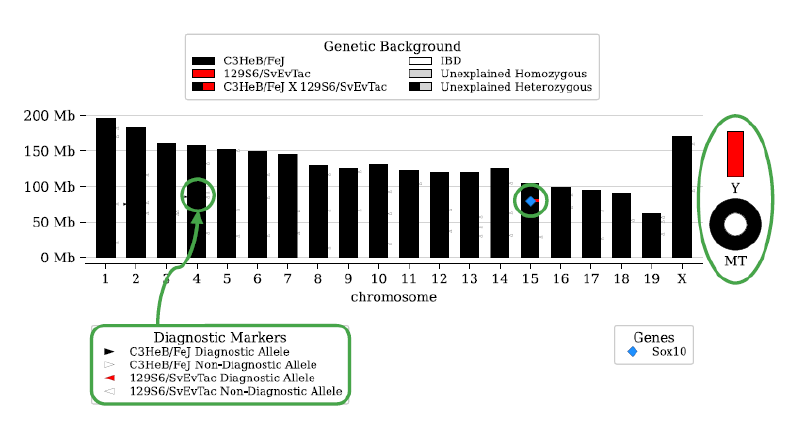
Dr. Pardo Manuel de Villena explained that questions on the use of vertebrate animals in research always are at the forefront of public and scientific discourse. Scientists face pressure to justify the use of animals, and standards for rigor and reproducibility are crucial to this argument. MiniMUGA offers a new tool to address this challenge (Figure 2).
“There is this questioning about the use of vertebrate animals in biomedical research. That is a question that, from my point of view, should always be asked. We need to reexamine how and why we do things,” Dr. Pardo Manuel de Villena reflected. “One of those things is asking: Is the genetics important? And to answer that, you need to have [quality control] over strain genetics.”
MiniMUGA: A Practical and Accessible Tool for Research Using Mice
MiniMUGA provides researchers a detailed understanding of the genetic background of their research specimens. The MiniMUGA array provides two key advantages over previous platforms used for this purpose: it is simple for non-experts to use, and it is available at low cost.
“[MiniMUGA] is intuitive, and that’s something of which I’m proud. It’s not only a great tool, but it’s an easy-to-use tool.” Dr. Pardo Manuel de Villena emphasized. He likened MiniMUGA to a hammer that strikes the nails of genetic ambiguity into place. “We are not looking for nails; there are nails all over the place,” Dr. Pardo Manuel de Villena said. “Now we can go and hammer those nails down and get these sorted out.”
MiniMUGA is an array-based platform that contains more than 11,000 probes—designed for either biallelic single-nucleotide polymorphisms (SNPs) (i.e., variations at single base pairs) or the presence of genetic constructs (i.e., inserted gene sequences)—to differentiate various laboratory mouse strains. The tool provides five unique features: (1) chromosomal sex determination, (2) discrimination between sub-strains from multiple commercial vendors, (3) diagnostic SNPs for popular laboratory strains, (4) detection of constructs used in genetically engineered mice, and (5) an easy-to-interpret report summarizing these results.1
Dr. Laura Reinholdt, Associate Professor at The Jackson Laboratory (JAX) and Co-Director of the JAX Mutant Mouse Resource and Research Center (MMRRC), explained that MiniMUGA provides an alternative to whole-genome sequencing, which generally is not feasible for researchers to perform on every specimen. She noted, however, that whole-genome sequencing could be informative in certain contexts (e.g., for studies involving engineered alleles). In these cases, nanopore sequencing offers additional capabilities. Additionally, targeted locus amplification is valuable for transgenic strains in which the genomic insertion site is unknown.
“We have so much more genetic information now,” Dr. Reinholdt stated. “It’s a little bit scary because when you look under the hood, you might find things that you didn’t expect. But it will make us better; it will make our strains better. … To me it seems if the technology is available, we have to use it.”
MMRRC: A Steward of Community Resources
The Office of Research Infrastructure Programs (ORIP)–funded MMRRC Program is at the forefront of developing and implementing novel broad-based strategies to optimize rigor and reproducibility in research using mouse models. This is exemplified by support of the development, testing, refinement, and application of MiniMUGA for the assessment and confirmation of background strain or sub-strain of mutant mice. Genetic background information and related services are available to investigators submitting and requesting strains from the MMRRCs (Figure 3).
The MMRRC Consortium maintains thousands of mouse strains, many of which have been housed at the repository for decades. Genetic quality control—for genetic backgrounds, as well as engineered alleles—is crucial to the Consortium’s mission. The challenge, Dr. Reinholdt stated, is to convey the importance of genetic quality control across the broad community that the MMRRC Consortium serves.
“The onus now is on us, as users and stewards of these strains, to make sure that people understand the information and understand how it’s important for their research, and [how it is interpreted], so people can [rely on] what’s been published,” Dr. Reinholdt said.
Dr. Reinholdt emphasized that adoption of genetic quality control standards represents a crucial challenge within scientific research. Although the Centers have played a leading role in conveying this message to the scientific community, investigators hold a responsibility to ensure that their work is documented properly, even prior to publication. Scientific journals and reviewers also must hold investigators to high standards by setting requirements that genetic backgrounds be documented properly prior to publication or grant award.
A Community-Wide Shift
Dr. Kent Lloyd, Professor in the Department of Surgery at the University of California, Davis (UC-Davis), and Director of the UC Davis Mouse Biology Program, reflected that reproducibility represents a longstanding issue within the scientific community: many scientists lack the training, background, and time to ponder the importance of genetic backgrounds. The key to the issue, Dr. Lloyd emphasized, lies in the training of young scientists. He underscored the importance of experimental design, methodology, honesty, transparency, and reproducibility. In a world in which the sheer volume of high-impact publications often is prioritized, Dr. Lloyd urged investigators to take time to ponder the implications of their work.
“I’m in that wheel, too—running, trying to keep up,” Dr. Lloyd reflected, likening himself to the mice that are central to his research. “And I don’t have enough time to think. And boy, if there’s any group of people that needs to think, it’s those who have been given the blessing of being able to have a scientific career.”
References
1 Sigmon JS, Blanchard MW, Baric RS, et al. Content and performance of the MiniMUGA genotyping array: a new tool to improve rigor and reproducibility in mouse research. Genetics. 2020;216(4):905–930. doi:10.1534/genetics.120.303596.
2 Amos-Landgraf J, Franklin C, Godfrey V, et al. The Mutant Mouse Resource and Research Center (MMRRC): the NIH‑supported national public repository and distribution archive of mutant mouse models in the USA. Mamm Genome. 2021;1–10. doi:10.1007/s00335-021-09894-0.



