Dr. Frank Slack, Director of the Harvard Medical School Initiative for RNA Medicine (HIRM), envisions a promising future for RNA-based therapeutics—a future he believes will be supported by core facilities like the Precision RNA Medicine Core Facility (RNA Core) at Beth Israel Deaconess Medical Center (BIDMC), a teaching affiliate of Harvard Medical School. The RNA Core provides investigators at BIDMC and beyond with affordable access to the latest RNA-based technology and bioinformatic expertise. According to Dr. Slack, who also serves as the Co-Director of the RNA Core, “The vision was that we would provide a concierge service where we could support a client—armed only with clinical samples—in moving from the discovery to the drug stage.”
The facility housing the RNA Core originally was built with ORIP funds (C06RR028581) administered through the 2009 American Recovery and Reinvestment Act (ARRA) grant program—whose goals were to stimulate the U.S. economy, create and preserve jobs, and advance scientific research. The grant was one of 148 ARRA awards that were granted through ORIP to support the construction or renovation of 64 animal facilities, 56 research laboratories, 17 clinical research facilities or laboratories, 8 imaging facilities, and 7 data centers. The award provided funds to BIDMC to convert a hospital basement laundry room into a state-of-the-art laboratory for onsite production of radioactively labeled tracers to support cancer research. With permission from the NIH, the space was repurposed to support the HIRM. The renovation helped the RNA Core secure a $3.5 million equipment grant from the Massachusetts Life Sciences Center (MLSC), a state agency with the mission of supporting life science growth and development in Massachusetts.
The RNA Core opened to clients in academia and industry at the end of 2018. Currently, it serves more than 50 clients per year and often processes more than 100 samples per month. The team (Figure 1) offers a full range of services via four main units: Delivery, Detection, Bioinformatics, and Spatial Technologies. The Delivery Unit generates nanoparticles for the packaging and delivery of RNA to cells and tissues. Researchers can request small-scale nanoparticle batches for preliminary trials in cell-based assays, then scale up their orders for larger preclinical trials in animal models. The Detection Unit houses several instruments that allow clients to detect, quantify, or visualize RNA in their samples (Figure 2).

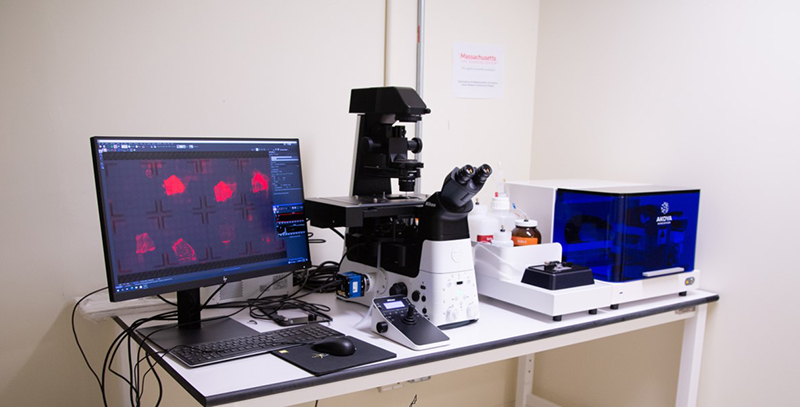
The equipment is sensitive enough to detect very low levels of RNA in such samples as blood or saliva, which are becoming new sources of biomarkers for several diseases. The Bioinformatics Unit is highly specialized for studying RNA, with an emphasis on RNA transcribed from noncoding DNA regions, which comprise about 98% of the human genome.1 “Our Bioinformatics Unit is trying to help people sift through RNAs being transcribed in cells and make sense of them,” explained Dr. Slack. “This could involve studying their expression levels, their structure, or whether the RNAs are mutated or not.”
The RNA Core has played an important role in supporting COVID-19 research efforts. “When the pandemic hit, our physician and nurse colleagues were working above and beyond to support our patients against this unknown and formidable foe. We decided to do what we do best and to contribute in this worldwide effort by characterizing how COVID-19 affects human organs and tissues at the cellular level,” explained Dr. Ioannis Vlachos, an Assistant Professor of Pathology at Harvard Medical School and the Director of the RNA Core Bioinformatics Unit at BIDMC. Bioinformaticians from the RNA Core worked with BIDMC pathologists and other experts to collect and analyze COVID-19 tissue samples, which sometimes involved visits to the hospital at 3:00 or 4:00 a.m. The team met almost daily via Zoom calls, spending hours discussing their results. These efforts resulted in a COVID-19 tissue atlas published in the journal Nature in April 2021.2 The paper described COVID-19 pathologies at the molecular and spatial levels, including altered RNA landscapes in the lungs and changes to the heart, kidney, and liver. Included in the paper were spatial transcriptomics data, where regions of interest on a single tissue slide—which were chosen using various biomarkers for SARS-CoV-2 infection—were analyzed for the expression of 18,000 genes. This technique combines in-depth transcriptomics data with spatial information that normally is lost when RNA is extracted from cells or tissues en masse.3 Following this successful initial effort and with additional support from the MLSC, the RNA Core established its Spatial Technologies Unit (STU) to provide access to single-cell and spatial tissue profiling instrumentation, technologies, and expertise to the community. The STU enables single-cell and in situ transcriptomics and multiomics, currently supporting researchers across the United States.
Researchers in the RNA Core are working with biotechnology companies to test and improve the next generation of RNA technologies. In the past year, much of the technology in the facility has been automated. The RNA Core is not yet clinically certified, but a goal for the future is to create potential new RNA-based or RNA-targeting drugs. “A silver lining of the pandemic has been the recognition of RNA medicine. It has been vindicating to those of us in the field who had a vision that one day we would be helping people this way,” explained Dr. Slack. As for the facility’s mission to increase access to RNA technology, both Dr. Slack and Dr. Vlachos noted growing interest among researchers in the field. “I believe that this is just the beginning of a paradigm shift, and we are in a very fortunate position to make the transition easy for others,” emphasized Dr. Vlachos.
Learn more information on the ARRA program and awards.
References
1 Piovesan A, Antonaros F, Vitale L, et al. Human protein-coding genes and gene feature statistics in 2019. BMC Res Notes. 2019;12:315. doi:10.1186/s13104-019-4343-8.
2 Delorey TM, Ziegler CGK, Heimberg G, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595:107–113. doi:10.1038/s41586-021-03570-8.
3 Rao A, Barkley D, França GS, et al. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596:211–220. doi:10.1038/s41586-021-03634-9.



