Micro-CT: A view into the structure of life for the analysis of tissue phenotypes
A convergence of scientific breakthroughs in chemistry, optics, and engineering in the early 1800s resulted in a miraculous new way to understand life—the light microscope. Microscopic studies of a wide range of samples by Johannes Müller and his protégés Matthias Schlieden, Theodor Schwann, and Rudolf Virchow resulted in a scientific epiphany and one of the most profound and useful revelations in history: Cells are the building blocks of all living organisms. Histology, the microscopic study of stained sections of fixed tissues, was to become a basis for a transformation of medicine at the dawn of the very next century.
Scientists now understand that morphological changes in cells and tissues occur with every major human disease, and these three-dimensional (3D) phenotypes can affect any cell type at any life stage. The detection of these phenotypes requires visualization of cellular features. Among today’s greatest scientific challenges is how to comprehensively and rapidly phenotype model organisms in large-scale efforts known as phenome projects. Yeast and mouse phenome projects were significant steps in this direction, but yeast is a single-cell organism and whole post-embryonic mouse specimens are too large to fully image in 3D at a cellular resolution. Pathologist and geneticist Dr. Keith Cheng at Penn State College of Medicine noted that the size of zebrafish makes them an ideal economical vertebrate model for eventual diagnostic computational analysis of human tissue specimens and for vertebrate phenomics involving high-throughput, comprehensive whole-organism tissue phenotyping of all cell types and organ systems.
In earlier work, the Cheng laboratory took advantage of histology to see submicron-scale cellular change across whole zebrafish1, 2 but was frustrated by their inability to reveal the larger millimeter-scale changes in vessels, nerve tracts, and skeleton3. Ideally, comprehensive, multiscale phenotyping would detect changes in the shape and size of both larger scale structures, such as skeletons or organs, as well as the smaller scale cellular features, including those characterizing cancer. Today, anatomic pathologists use cytological characteristics in descriptive, qualitative tissue assessments based on only 1 to 2 percent of tissue samples. Using full tissue samples to quantify the geometry of cellular changes would diminish sampling error and greatly increase precision in tissue diagnostics. Dr. Cheng decided to take a systems approach to addressing these issues.
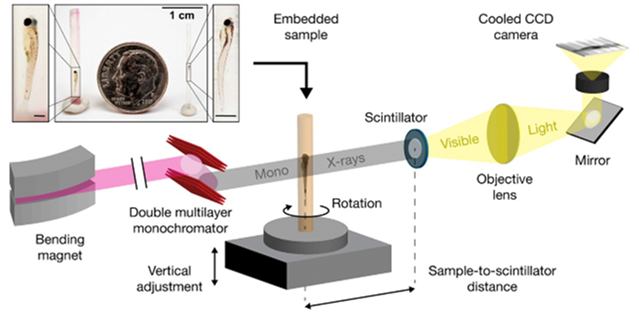
The basis of the approach proposed by Dr. Cheng with physicist Dr. Patrick La Rivière at the University of Chicago and colleagues at Argonne National Laboratory was X-ray microtomography (micro-CT). Micro-CT uses the same principles as human computed tomography scans but can achieve 1,000-fold higher resolution. The team worked for more than a decade to solve an array of technical problems and challenges to build a 3D mechanism for accurate quantification of histological features across all cell types of millimeter- to centimeter-scale whole specimens, including sample preparation, imaging physics, image processing, and sharing of massive data files. This advance was made possible by funding of a R24 grant (R24OD018559) from NIH’s Office of Research Infrastructure and Programs (ORIP).
For pathologists to detect and characterize each individual cell in a whole vertebrate organism, the imaging technology needed to achieve a combination of field of view, tissue contrast, and resolution. Further advancements in imaging speed were needed to overcome the handling of tens of thousands of samples involved in the phenome projects. Commercial sources used in Dr. Cheng’s initial work were slow and had fields of view that were too small to image whole zebrafish. Only synchrotrons (particle accelerators that use electric and magnetic fields to impart high speeds to charged particles and serve as a powerful source of x-rays) have adequate flux for high-speed imaging. The team then performed the necessary groundwork to create a new synchrotron-based imaging resource for biology, working with physicists at the Advanced Photon Source synchrotron at the Argonne National Laboratory (Fig.1). Their recently published results4 show that they are a step closer to the reality of high-throughput and multiscale phenotyping of cell and tissue change in an intact, whole vertebrate organism that is similar in size to human tissue specimens.
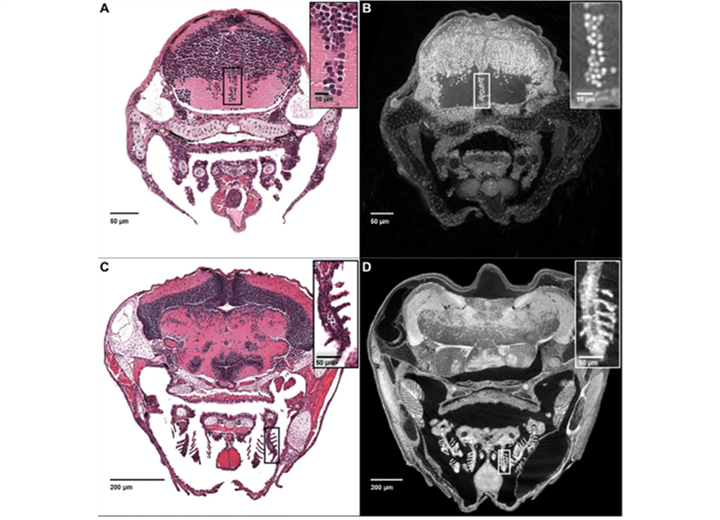
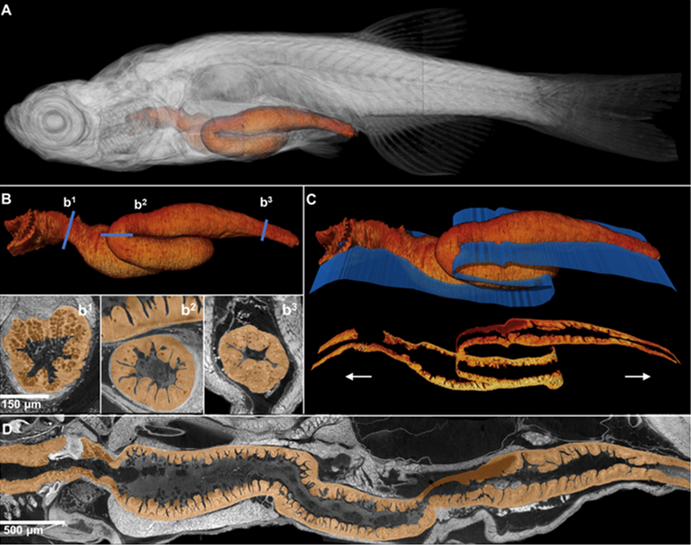
Optimizations of sample preparation and scanning parameters allowed the team to image entire larval and even juvenile zebrafish at about 1 micron3 resolution to achieve histology-like virtual sections that are computationally generated (Fig. 2). Because of the inherent isotropic nature of these reconstructions—that is, equal in scale in each dimension—virtual sections at histological resolutions can be generated at any angle, allowing scientists to sample structures in or along any continuous linear or curved plane (see juvenile fish gastrointestinal tract, Fig. 3). Because of the combination of field of view and resolution, other unique visualizations of both large-scale structure and cellular detail are now possible (visualization of both macro- and microanatomy shown in Movie 1 and Movie 2). The laboratory also created web tools for users to interact with the huge data sets (100 gigabytes per fish) at ViewTool (www.3d.fish).
At submicron resolutions, it becomes possible to measure the shape and volume of cells and nuclei. Machine learning automates these measurements. By computationally locating the centers of each nucleus of the brain, Cheng’s team was able to compute the number of cells within a chosen volume of the fish, which led to a computation of cell density4. To the researchers’ surprise, the brains of wild-type sibling larvae can have distinct phenotypic differences; these variations are important to understand the physiology of disease (Fig. 4).
The team is further growing the micro-CT technology to pursue large-scale disease model validation and phenome projects. Dr. Cheng has set the goal of increasing imaging speed to one sample per minute. The team is pursuing improvements in field of view and resolution and is developing machine-learning techniques to automate the recognition, quantification, and modeling of anatomical structures and phenotypes. Dr. Cheng has set a resolution benchmark at 0.5 microns to achieve an exciting goal: the visualization of every condensed mitotic chromosome in a whole vertebrate organism.
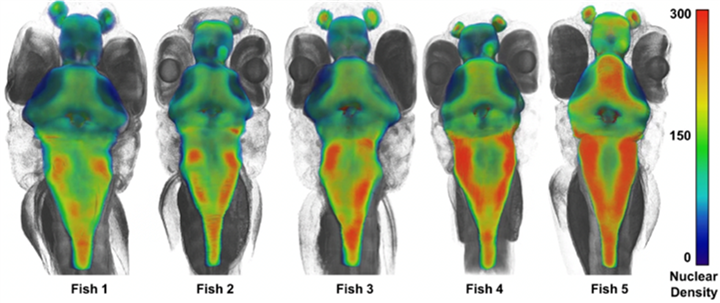
Figure 4. Analysis of cell density in the brains of wild-type sibling larvae demonstrated striking variations between brain regions and between siblings.
The broader, longer term vision includes the creation of a large-scale, dedicated synchrotron-based imaging resource for biology. This ambitious goal would generate petabytes of data for the entire biomedical research community to explore applications for environmental toxicology, drug discovery, and disease diagnostics, among others. A network of synchrotron beamlines would enable the once-impossible goal of high-throughput, standardized whole-organism imaging for multi-investigator phenome projects. The application of artificial intelligence and robotics to this network could enable Dr. Cheng’s team to begin to reach a tantalizing level of knowledge: defining a geometry of life and phenotype across all living things. This foundation would help them gain more comprehensive insight into the function of every vertebrate gene and the full range of change caused by every chemical, for every cell type and every organ system.
Acknowledgments
ORIP thanks Dr. Cheng and his laboratory for their significant contributions in developing this article.
References
1Mohideen MA, Beckwith LG, Tsao-Wu GS, Moore JL, Wong AC, Chinoy MR, Cheng KC. Histology-based screen for zebrafish mutants with abnormal cell differentiation. Dev Dyn. 2003; 228(3): 414-23.
2Cheng KC, Xin X, Clark DP, La Riviere P. Whole-animal imaging, gene function, and the Zebrafish Phenome Project. Curr Opin Genet Dev. 2011; 21(5): 620-9.
3Cheng KC, Katz SR, Lin AY, Xin X, Ding Y. Whole-Organism Cellular Pathology: A Systems Approach to Phenomics. Adv Genet. 2016;95: 89-115.
4Ding Y, Vanselow DJ, Yakovlev MA, Katz SR, Lin AY, Clark DP, Vargas P, Xin X, Copper JE, Canfield VA, Ang KC, Wang Y, Xiao X, De Carlo F, van Rossum DB, La Riviere P, Cheng KC. Computational 3D histological phenotyping of whole zebrafish by X-ray histotomography. eLife. 2019;8. pii: e44898.



