Gnotobiotic Mouse Resources Support Studies of Microbial Interactions
Microorganisms affect nearly every aspect of life, regulating numerous biological processes at the ecosystem and organismal levels. All multi-organ animals maintain a unique gut microbiome that plays a key role in regulation of their metabolic, immune, and neurological functions. This area of research has expanded thanks to studies catalyzed by the NIH Human Microbiome Project, but the functional consequences of microbial changes—and the role of individual bacterial species and combinations—remain unknown. The field of gnotobiology was established to help investigators address this concern in their studies, providing a better understanding of microbe–microbe and host–microbe interactions using animal models in whole-animal contexts (Figure 1).1,2
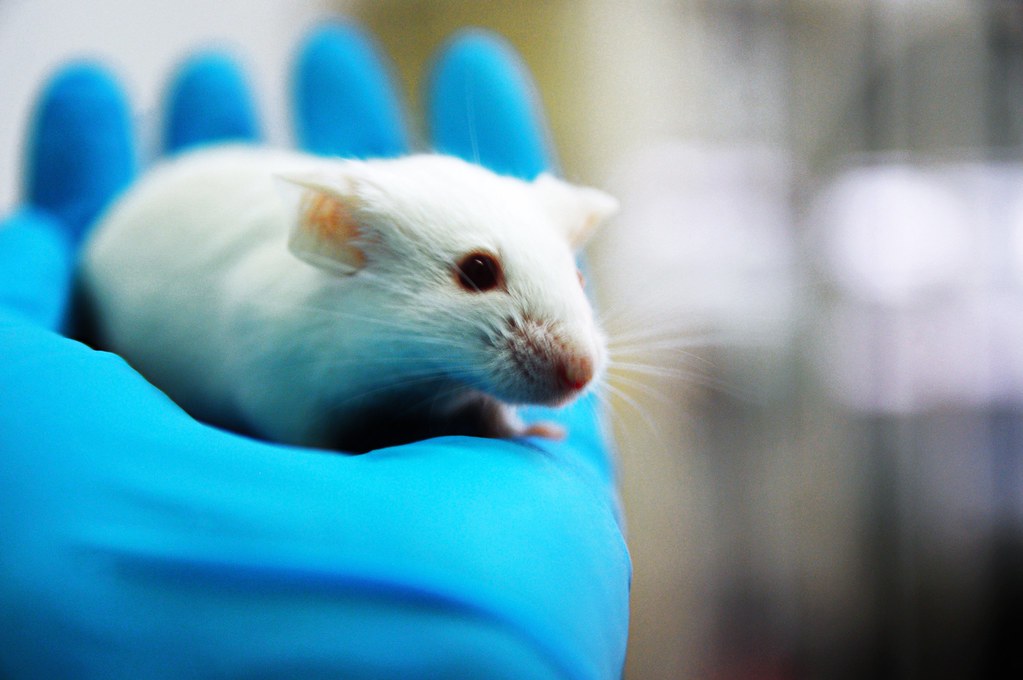
Originating from the Greek “gnōtos” (i.e., known) and “bios” (i.e., life), gnotobiology refers to organisms that contain only known microorganisms, permitting studies in a controlled biologic environment for research. Gnotobiology enables precise microbial manipulation by colonizing germ-free (i.e., sterile) mice (or other animal species) with single or combined resident or pathogenic bacterial, viral, or fungal species, including genetically engineered bacterial strains. Alternatively, gnotobiotic animals can be colonized via transplantation of complex fecal bacterial communities derived from normal or diseased humans or experimental animals.
“You can look at the big picture—including every bacterium, virus, [and] fungus—which becomes overwhelming. Or you can break it down and look at the function of a single bacterium,” stated Dr. R. Balfour Sartor, the Margaret and Lorimer W. Midget Distinguished Professor of Medicine at The University of North Carolina (UNC) at Chapel Hill School of Medicine. “If I see that one bacterium causes inflammation, I may hypothesize that it’s doing it by a particular mechanism. You can investigate that mechanism in cell culture, but that does not consider the interactions of the in vivo complex environment—mucus layers, epithelial barrier, different types of immune cells—all interacting with each other. That’s almost impossible to reproduce in an in vitro study.”
To better understand the dynamics of microbial interactions in vivo, investigators can make use of gnotobiotic animals for research. ORIP is committed to funding gnotobiotic resources for the biomedical research community. Established through ORIP support in 2004, the National Gnotobiotic Rodent Resource Center (NGRRC) at UNC provides germ-free and gnotobiotic mice to NIH-funded researchers across the United States. The NGRRC enables investigators to explore the broad hypothesis that resident bacteria are protective in normal hosts but induce pathogenic inflammatory, metabolic, and neoplastic responses in susceptible hosts with genetic risks or environmental triggers. Dr. Sartor, who serves as NGRRC Director, explained that the Center provides a valuable resource to the biomedical research community.
“Developing, deriving, and maintaining a mouse in the absence of any detectable [microbiota] is an expensive and time-consuming task beyond the reach of most individual investigators. And yet these mice are extremely valuable,” Dr. Sartor emphasized. “Our Center provides a convenient and cost-effective resource for investigators around the country to study whatever bacteria, virus, or fungi in whatever organ of interest or disease process they’re interested in.”
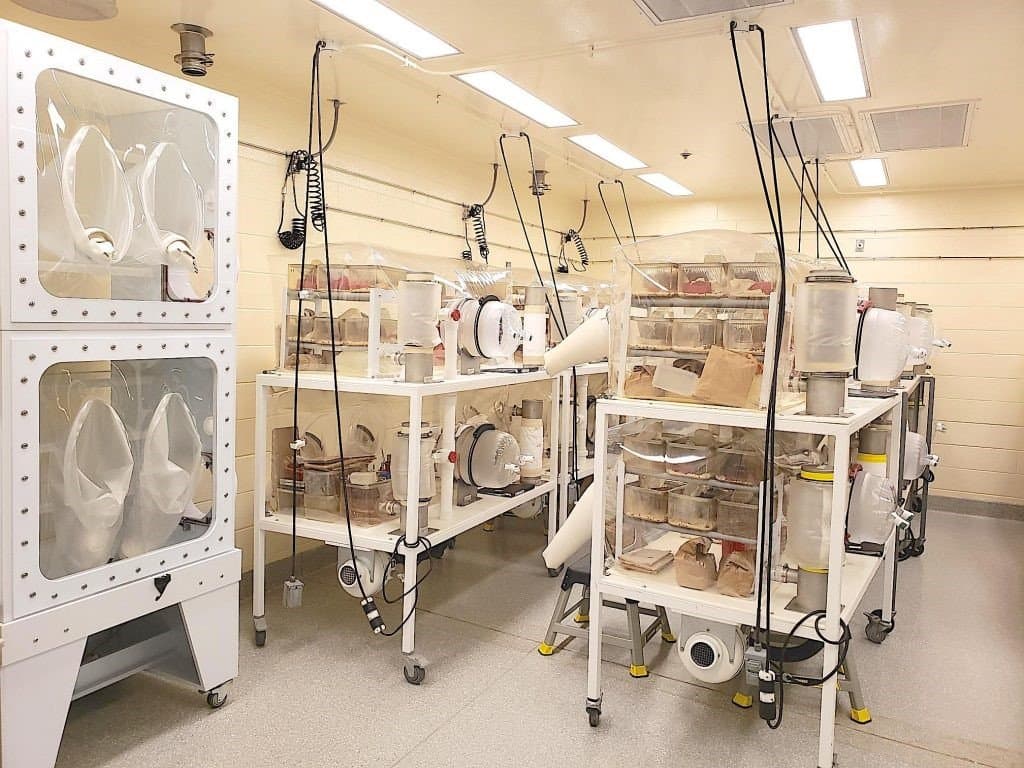
The NGRRC maintains about 30 strains of mice within a germ-free environment, providing a variety of genetic backgrounds that researchers can select for their studies. The colonies are maintained in individual isolator units that allow staff members to perform husbandry while preventing contamination (Figure 2). To create gnotobiotic populations, the animals are colonized with a known microbiota—or combinations of microbiota—to characterize their functions in various biological contexts. NGRRC’s expert staff colonizes the mice with bacterial strains, performs experiments, and harvests biological samples for investigators who lack the capability for germ-free facilities at their own institutions.
Investigators also can visit the NGRRC to undergo a hands-on training course, which provides them with the knowledge and skills needed to establish gnotobiotic facilities at their home institutions. The NGRRC has assisted more than 100 NIH investigators—both intramural and extramural—from 52 institutions, and the Center’s services span multiple areas of biomedical research representing at least seven NIH Institutes and Centers.
NGRRC’s efforts are helping to promote increased rigor and reproducibility of biomedical research. Because the microbiome can modify the phenotype of the model organisms, it is vital to study the role of the microbiome in diseases, monitor husbandry, search for critical extrinsic factors affecting research studies, and develop recommendations and new tools for the research community.
Dr. Sartor, whose research is also funded by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), studies the dynamics of the gut microbiome in intestinal inflammation. His team has developed a humanized mouse model—using fecal transplants from human patients—to study how commonly used Western dietary additives potentiate colitis and to validate and refine a protective consortium of rationally designed human bacteria.3,4 Another example of a researcher who has leveraged the NGRCC is Dr. Jasmohan Bajaj, Professor in the Department of Internal Medicine at the Virginia Commonwealth University School of Medicine. Dr. Bajaj has collaborated with the NGRRC to perform studies of brain and liver function in gnotobiotic mice using fecal transplants from human patients with alcohol‑induced cirrhosis.5 His group is developing inflammatory profiles that provide insight on the use of fecal transplants to reduce alcohol cravings. Dr. Bajaj has received funding from the NIDDK and the National Center for Advancing Translational Sciences to perform this work.
Dr. Sartor emphasized that the NGRRC has been critical to numerous areas of research; its staff are developing innovative approaches continuously to meet investigators’ needs. ORIP’s support has allowed the Center to provide its services to a large number of investigators at a fraction of the commercial cost. “ORIP’s support has been absolutely essential to allow investigators from many different disciplines to do cost-effective work,” he emphasized. “It’s absolutely necessary for us to provide this resource for other investigators in the research community.”
For more information on ORIP’s support for the NGRRC and other rodent resources, please download ORIP’s Rodent Resources Fact Sheet.
References
1 Martín R, Bermúdez-Humarán LG, Langella P. Gnotobiotic rodents: an in vivo model for the study of microbe–microbe interactions. Front Microbiol. 2016;7:409. doi:10.3389/fmicb.2016.00409.
2 Rogala AR, Oka A, Sartor RB. Strategies to dissect host–microbial immune interactions that determine mucosal homeostasis vs. intestinal inflammation in gnotobiotic mice. Front Immunol. 2020;11:214. doi:10.3389/fimmu.2020.00214.
3 Rousta E, Oka A, Liu B, et al. The emulsifier carboxymethylcellulose induces more aggressive colitis in humanized mice with inflammatory bowel disease microbiota than polysorbate-80. Nutrients. 2021;13(10):3565. doi:10.3390/nu13103565.
4 van der Lelie D, Oka A, Taghavi S, et al. Rationally designed bacterial consortia to treat chronic immune-mediated colitis and restore intestinal homeostasis. Nat Commun. 12(1):3105. doi:10.1038/s41467-021-23460-x.
5 Liu R, Kang JD, Sartor RB, et al. Neuroinflammation in murine cirrhosis is dependent on the gut microbiome and is attenuated by fecal transplant. Hepatology. 2020;71(2):611–626. doi:10.1002/hep.30827.



