Cryo-Electron Microscopy Used to Bridge the Micro-Nano Gap
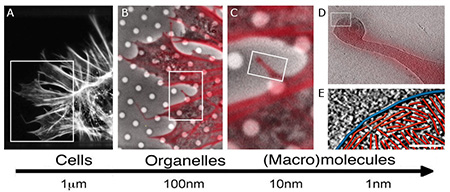
To understand human physiology and the pathology of diseases, it is important to investigate the underlying biological processes on all spatial scales. These scales range from the patient to organs, to tissues within an organ, to individual cells within the tissue, to molecular machines within the cells, down to the atomic level. The detailed knowledge of macromolecular interactions within and in between cells in a given tissue, combined with an integrated view of an organism, significantly increases the chances for finding new cures.
Since its invention in the 1930s by Ernst Ruska and Max Knoll, the electron microscope has been used to capture images of biological specimens. The technological advances of the 1940s allowed the observation of cellular organelles embedded in heavy metal stains by transmission electron microscopy (TEM). The 1960s brought the ability to generate 3D structures from individual macromolecular assemblies. Development of cryogenic electron microscopy (cryoEM) in the 1970s provided the means to image these macromolecules while keeping them hydrated in biochemically functional buffer conditions by instantaneously freezing them at −180°C. However, the resolution attainable for these samples through cryoEM was limited. At that time, therefore, X-ray crystallography was the method of choice for resolving molecular structures. As a robust and mature technology, X-ray crystallography continues to provide de novo structural models, but it is not without limitations. For example, X-ray crystallography requires isolation and crystallization of large quantities of the molecules, and these processes often are difficult and time-consuming. Also, the resulting crystals may capture the molecules in a conformation that does not occur naturally in biologically relevant environments. Last, molecules of high medical importance often are very hard or practically impossible to crystallize.
Recent advances in hardware and image analysis capabilities have revolutionized the field of cryoEM, enabling the generation of high-resolution structural data from vitrified samples at resolutions that rival X-ray crystallography, without the need for crystallization. In addition, cryoEM is parsimonious in its material requirements and is capable of handling fully hydrated pleiomorphic macromolecular assemblies. Currently, one of the most powerful cryoEM instruments is the FEI Titan Krios. Titan Krios’ greatest advantage over earlier instruments is its robust data collection capability. The instrument can generate large volumes of high-resolution, high-contrast data from automated data collection runs, typically within 3-6 hours. The analysis of these data yields molecular structures with the resolution of a fraction of a nanometer (2–3Å) that rivals resolution of structures obtained with X-ray diffraction methods.

The National Institutes of Health (NIH) Office of Research Infrastructure Programs (ORIP), through its Shared Instrumentation Program (S10), supports grant awards for the acquisition of cutting-edge instruments that enable and enhance research of NIH-supported investigators. ORIP awarded an S10 grant to the Sanford Burnham Prebys (SBP) Medical Discovery Institute to support the purchase of the Titan Krios (Figure 1), commissioned in 2015. A separate NIH shared instrumentation award to SBP in 2008 funded ancillary instruments required for the preparation of samples for cryoEM experiments. Setting up the cryoEM research program at SBP also required dedicated high-end computers, adequate data storage capacity, and sophisticated software for on-the-fly processing and real-time feedback on the quality of the data collected—all serving to optimize microscope usage. The cryoEM research program also required additional investments, such as the allocation of space, maintenance, and support for highly trained personnel to operate the instruments.
The availability of the Titan Krios and ancillary equipment at SBP, made possible through the S10 award, has enormously increased the Institute’s imaging capabilities for determining high-resolution macromolecular structures. For Dr. Dorit Hanein, a former principal investigator at SBP (currently at Scintillon Institute), the technology offered an opportunity for high-impact, rigorous, and robust discoveries in basic biological research and medicine in situ by permitting the investigation of macromolecules inside mammalian cells at high resolution. Over the years, the instrument also supported collaborative projects between researchers at SBP and many institutions nationwide, including The University of North Carolina Chapel Hill, Yale University, and Los Alamos National Laboratory.
Researchers at SBP together with their collaborators the developed experimental workflows based on integrating imaging modalities—such as correlative fluorescence light microscopy, cellular cryo tomography, and cryoEM—across spatial and temporal scales (Figure 2) to investigate biological processes.1–8 This approach, when combined with image analysis and data mining, allows investigators to identify the location of components of macromolecular complexes in a whole cell and provide information about their function.
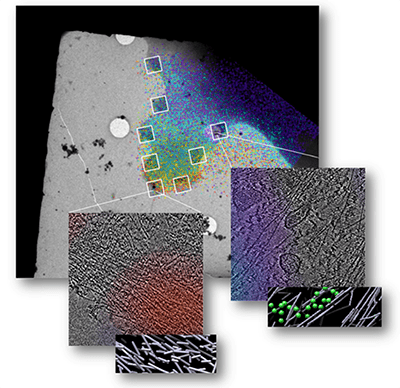
Within the last 2 years, investigators at the SBP have completed several research studies using the Titan Krios instrument. Together with Drs. Klaus Hanh, Daniel J. Marston, and others at The University of North Carolina at Chapel Hill, Drs. Hanein and Neils Volkmann and their colleagues at SBP combined cellular cryo-tomography (cryoET) and the Förster resonance energy transfer (FRET) light microscopy technique to identify underlying structural morphologies associated with states of activation of Rho GTPases, a family of proteins that regulate the dynamics of actin within the cell.9 One member of this family, Rac1, is known to be a regulator of the actin cytoskeleton and generates significant morphological changes in the cell. In this study, the researchers observed the state of activation of Rac1 in real time and studied the underlying structure of actin at each stage of activation. They found that when Rac1 activity is high, a unique, spatially localized, 3D stack of very short, non directional actin filaments is generated (Figure 3). The researchers posit that this stack serves as a scaffolding system that can very quickly transition to the next structural elements the cell needs—for example, to generate a protrusion. The workflow the researchers developed using the Titan Krios can serve as a tool to study the dynamics of this scaffolding system and other macromolecular machines.
Another study, conducted by investigators at SBP and colleagues at Yale University, set out to investigate how mechanotransduction of force is manifested at the molecular level, and how it affects the structural organization of the actin cytoskeleton.10 The investigators examined talin, a protein that is critical for force transmission between the actin cytoskeleton and the extracellular matrix as well as being important for cells sensing the mechanical properties of their environment. Using a previously characterized FRET-based talin tension sensor, the researchers were able to determine a spatiotemporal relationship between talin tension, actin localization, local actin organization, and focal adhesion dynamics. They used this information about variations in tension to study the organization of the cytoskeleton in the cell at nanometer resolution.
Investigators at SBP and the Los Alamos National Laboratory also used the Titan Krios to study full-length, membrane-embedded molecules. The investigators had been seeking to understand, at the level of the membrane environment, how proteins cooperate with each other to induce cell death. By correlating technologies—nuclear magnetic resonance, molecular dynamic simulations, and the Titan Krios’ cellular cryoET—the researchers have been able to develop a more complete picture of the membrane-embedded protein BAX, which plays a pivotal role in triggering cell death.11
The 2017 Nobel Prize in Chemistry went to three investigators for the development of cryoEM techniques that simplify and improve the imaging of biomolecules, which has given the field a significant boost.
References
1 Page C, Hanein D, Volkmann N. Accurate membrane tracing in three-dimensional reconstructions from electron cryotomography data. Ultramicroscopy. 2015;155:20–26.
2 Suraneni P, Rubinstein B, Unruh JR, et al. The Arp2/3 complex is required for lamellipodia extension and directional fibroblast cell migration. J Cell Biol. 2012;197:239–251.
3 Suraneni P, Fogelson B, Rubinstein B, et al. A mechanism of leading-edge protrusion in the absence of Arp2/3 complex. Mol Biol Cell. 2015;26(5):901–912.
4 Anderson KL, Page C, Swift MF, et al. Nano-scale actin-network characterization of fibroblast cells lacking functional Arp2/3 complex. J Struct Biol. 2017;197(3):312–321.
5 Xu XP, Kim E, Swift M, et al. Three-dimensional structures of full-length, membrane-embedded human α(IIb)β(3) integrin complexes. Biophys J. 2016;110(4):798–809.
6 Chen Z, Sun L, Zhang Z, et al. Cryo-EM structure of the bacteriophage T4 isometric head at 3.3-Å resolution and its relevance to the assembly of icosahedral viruses. Proc Natl Acad Sci U S A. 2017;114(39):E8184–E8193.
7 Anderson KL, Page C, Swift MF, et al. Marker-free method for accurate alignment between correlated light, cryo-light, and electron cryo-microscopy data using sample support features. J Struct Biol. 2018;201(1):46–51.
8 Buckley CD, Tan J, Anderson KL, et al. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 2014;346(6209):1254211.
9 Marston DJ, Anderson KL, Swift MF, et al. High Rac1 activity is functionally translated into cytosolic structures with unique nanoscale cytoskeletal architecture. Proc Natl Acad Sci U S A. 2019;116(4);1267–1272.
10 Kumar A, Anderson KL, Swift MF, et al. Local tension on talin in focal adhesions correlates with F-actin alignment at the nanometer scale. Biophys J. 2018;115:1569–1579.
11 Lopez CA, Swift MF, Xu X, et al. Biophysical characterization of a nanodisc with and without BAX: An integrative study using molecular dynamics simulations and cryo-EM. Structure. 2019;27(6):988–999.



