The Center for Precision Animal Modeling at The University of Alabama at Birmingham Advances Disease Models for Research
In recent years, the research community has become increasingly interested in precision disease modeling, and it remains at the forefront of personalized medicine approaches for developing models for human disease. In 2014, ORIP launched the Precision Disease Modeling Initiative (PAR-14-280) and subsequently funded three Pilot Centers for Disease Modeling (U54) in 2015. Given the success of the initial cohort (see Archives), ORIP renewed this initiative in 2020 (PAR-20-085) and awarded three new U54 centers. The University of Alabama at Birmingham (UAB) Center for Precision Animal Modeling (C-PAM) is one of the three centers established to provide the biomedical community with advanced animal models needed to develop precision therapies for monogenic and complex disorders, such as ciliopathies and Roberts syndrome.
While variants and genes appear to be linked to patient symptoms, the evidence of causality is often insufficient, leaving patients, families, and physicians who care for the patients without a definitive solution. Accordingly, ORIP revised its Precision Disease Modeling program to increase its focus on creating pipelines for research community–nominated unique human genomic variants linked to diseases for cost-effective, high-throughput testing in such models as mouse, Caenorhabditis elegans, zebrafish, and human induced pluripotent stem cells (iPSCs).
Dr. Bradley K. Yoder, Professor and Chair of Cell, Developmental, and Integrative Biology (CDIB), UAB (Figure 1), and Dr. Matthew Might, Director of the Hugh Kaul Precision Medicine Institute, Endowed Chair of Personalized Medicine, and Professor of Internal Medicine and Computer Science, UAB (Figure 2), are Director and Co-director of C-PAM, respectively.

They were awarded a 5-year grant (U54OD030167) from ORIP in 2020 and have led C-PAM in its mission to provide a national service to generate precision disease animal models for researchers and physicians working to diagnose, treat, and study genetic diseases. Dr. Yoder explained that C-PAM’s precision disease models—which are developed in iPSCs, worms, zebrafish, frogs, mice, and rats—are engineered to have the exact patient-specific variants known to be behind these diseases. He remarked, “UAB had limited infrastructure to support this research, and C‑PAM started from the ground up.” An interest group composed of 20 UAB Precision Medicine Institute members (clinicians, computational biologists, animal modelers, geneticists, and cell biology investigators) spearheaded this effort. The group meets weekly to discuss patient variants submitted to the center for analysis and to prioritize those that would benefit most from the generation of an animal or cell-based model. The threefold aim is to understand the mechanisms of each disease, identify potential treatments that will help patients with rare diseases live fuller lives, and fill the gap for rare disease patient advocacy groups (PAGs) at foundations that have identified variants in genes. Common databases, such as the Clinical Genome Resource (known as ClinGen), do not have information about these gene variants as causative, pathogenic changes. Many classified as variants of unknown significance. C‑PAM’s goal is to help bridge the gap between those patients and a final diagnosis.

C-PAM consists of five integrated sections—the Bioinformatics Section, Coordination Section, Disease Modeling Unit, Pre/Co-Clinical Section, and Resource and Services Section—that are essential for the efficient generation and analysis of precise animal models of direct relevance to human disease. Each section comprises multidisciplinary researchers who work to fulfill the mission of C-PAM. The aims are to assess the variants using computational approaches, generate pathogenicity predictions, prioritize variants and select those for modeling, and generate an animal or cell model for disease, primarily using CRISPR-Cas9 engineering. C-PAM’s userbase is composed of clinicians, foundation centers, patients, and PAGs.
One strength of C-PAM is the many animal species and mammalian cells it uses and their value for drug screening. Dr. Yoder noted that the goal is to mimic the human disease, find new gene variants in patients, develop animal models, and disseminate those models to interested investigators to inform and advance future studies on disease mechanisms and possible treatments. To facilitate this process, the Bioinformatics Core developed a tool called Rosalution,1 which is a framework for gathering all public information about a gene variant and its interactions. Rosalution streamlined the weekly meetings for reviewing variants from hours to minutes, providing a platform for gathering and sharing all of the data from each case. Rosalution has been made available to the public and can be accessed through GitHub. In addition, the bioinformatics team is using an artificial intelligence system, mediKanren, to search for compounds that may interface with this gene (upstream or downstream) as a potential therapeutic candidate.2 Predictions are made on repurposed drugs (already tested in clinical trials) that could interface with a variant to speed the results to patients. In the next step of the process, one or more animal models are developed.
An additional advantage of C-PAM is its use of Xenopus models to perform rapid pathogenicity prescreens, with a 1-week turnaround on the results. Clinicians commonly request Xenopus pathogenicity prescreens to rapidly prioritize multiple variants identified in their patients, and these requests are independent of animal model development. Two or more variants can be identified in a patient, screened, and overexpressed in Xenopus by injecting the RNA into a Xenopus egg. The response informs whether any patient variant has an altered function compared with the wild type and helps support the more costly and time-intensive animal model development and analysis.
Another key feature is that C-PAM generates comparative models across multiple species, with the goal of performing drug screens, which are more relevant to the patient than pathological assessments alone. Once the model is produced, the team performs basic phenotyping. Dr. Yoder remarked, “Phenotyping has been one of the bottlenecks because it is labor intensive, slow, and expensive.” The desired outcome is to define the phenotypic changes in the animal models that closely mimic what occurs in the patient, along with any unexpected phenotypes that were not originally identified in the clinical report. C-PAM reports findings to the physician caring for the patient or a foundation and asks whether they have checked for this new phenotype. In an iterative process, the patient information indicates how to improve the model and future care for the patient. The animal models are delivered to the submitting investigator and made available to other interested investigators for follow-up studies.
An additional strength of C-PAM has been establishing collaborations to facilitate dissemination of the animal models to the public for further analysis. Since its inception, the C-PAM network has received 164 disease variant nominations, accepted 87 nominations, and developed 118 approved models. The models span a wide range of disorders and phenotypes across all major categories of human genetic disease. In 2023, the decision was made to stop accepting new variant nominations to address the backlog and finish this funding cycle. The center expects to reopen the variant submission pipeline toward the end of 2024.
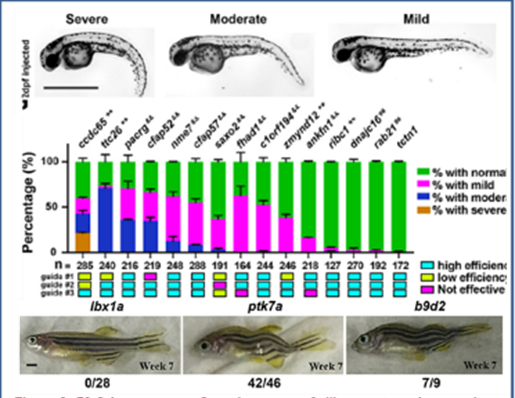
Over the course of the program, C-PAM has had several accomplishments and revealed many insights into variant pathogenicity. As an example, it identified variable phenotypes and penetrance (Figure 3) between and within different zebrafish ciliary transition zone mutants.3
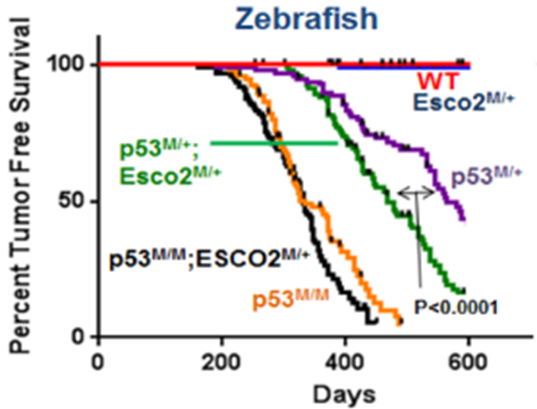
Researchers discovered that reduced sister chromatid cohesion acts as a tumor penetrance modifier in a zebrafish model for Roberts syndrome (Figure 4), which is a developmental disorder that has been linked to recessive inactivation of the ESCO2 (establishment of sister chromatid cohesion N-acetyltransferase 2) gene.4
C-PAM researchers also increased the understanding of tissue- and cell-specific phenotypes in SET binding protein 1 (SETBP1)–associated diseases, including the ultrarare Schinzel Giedion syndrome and SETBP1 haploinsufficiency disorder in mouse5 and human tissues.6 All C-PAM codes are open source and can be accessed through GitHub.
Dr. Matthew Alexander, one of C-PAM’s Resource and Services Section leaders, noted a major success of this program. A family whose son had an undiagnosed muscle disorder visited a neuromuscular specialist at Children’s of Alabama hospital, which submitted the case to C-PAM. The entire family had their genomes sequenced, and a previously unknown genetic variant in the vacuolar ATPase assembly factor (VMA21) gene was found. A diagnosis of X-linked myopathy with excessive autophagy (XMEA) was confirmed. The grandfather who had the disorder passed away in his 60s, and XEMA is now diagnosed in two of his cousins. Dr. Alexander explained that two XMEA animal models were developed, a drug screen was performed, and a key pathway was identified that appears to be affected in the C-PAM models. A manuscript is in progress in collaboration with Dr. James (Jim) Dowling at the Hospital for Sick Children (SickKids). This project highlights C-PAM’s full suite of services in the pipeline and the center’s collaborative efforts.
Additionally, outreach to increase awareness of the models has included presenting findings at national meetings and sponsoring booths at genetic scientific meetings. In terms of cross-center interactions, the three U54-funded Pilot Centers for Disease Modeling (UAB C-PAM, Baylor College of Medicine Center for Precision Medicine Models, and The Jackson Laboratory Center for Precision Genetics) meet quarterly to strategize on the best use of resources for projects submitted simultaneously to multiple centers. One center takes the lead in developing the model.
Dr. Yoder noted that it takes about a year to generate a rodent model. In 4 years, they built a team, integrated and established the pipeline, and advanced models to use in the clinical setting. He remarked, “We are just now at the phase where animals are starting to come out of our pipeline. C-PAM is right at that point where we could be truly impactful.” Continuing the C-PAM program will be beneficial to PAGs because they have a desire to find cures and treatments but do not have access to or fully understand the details of rare disease animal models and are not aware of the existing models. C-PAM fills this gap and provides the necessary connections for PAGs in a timely manner. C-PAM received a supplement to enhance high-throughput screening and phenotyping and will work with its collaborators in the next phase to perform drug screening. To further increase its impact, C-PAM leadership is exploring collaborations with the Advanced Research Projects Agency for Health (commonly called ARPA-H) and the UAB Center for Clinical and Translational Science to continue its efforts.
The Precision Disease Modeling Initiative is one of ORIP’s major projects. For more information, please visit ORIP’s Precision Disease Modeling website.
References
1 Uno-Antonison AE, Fatima R, Scherer J, et al. Rosalution: Supporting data accessibility, integration, curation, interoperability, and reuse for precision animal modeling. J Open Source Softw. 2023;8(90):5443. doi:10.21105/joss.05443.
2 Foksinska A, Crowder CM, Crouse AB, et al. The precision medicine process for treating rare disease using the artificial intelligence tool mediKanren. Front Artif Intell. 2022;5:910216. doi:10.3389/frai.2022.910216.
3 Wang J, Thomas HR, Thompson RG, et al. Variable phenotypes and penetrance between and within different zebrafish transition zone mutants. Dis Model Mech. 2022;15(12). doi:10.1242/dmm.049568.
4 Wang J, Thomas HR, Chen Y, et al. Reduced sister chromatid cohesion acts as a tumor penetrance modifier. PLOS Genet. 2022;18(8):e1010341. doi:10.1371/journal.pgen.1010341.
5 Whitlock JH, Soelter TM, Howton TC, et al. Cell-type-specific gene expression and regulation in the cerebral cortex and kidney of atypical Setbp1S858R Schinzel Giedion Syndrome mice. J Cell Mol Med. 2023;27(22):3565–3577. doi:10.1111/jcmm.18001.
6 Whitlock JH, Wilk EJ, Howton TC, et al. The landscape of SETBP1 gene expression and transcription factor activity across human tissues. PLoS One. 2024;19(1):e0296328. doi:10.1371/journal.pone.0296328.



