The Amazing Axolotl: A Valuable Model for Regenerative Medicine
Axolotls (Ambystoma mexicanum) are salamanders that have been used for research for more than 200 years (Figure 1).1 They have a remarkable ability to regenerate lost or damaged tissues, including whole organs, limbs, and parts of the central nervous system. Scientists study the genetic and biochemical mechanisms that drive axolotl tissue regeneration in hopes that deeper understanding may bridge the gap between regenerative biology and medicine.

Dr. Randal Voss, director of the Ambystoma Genetic Stock Center (AGSC) at the University of Kentucky (Figure 2), has studied axolotl evolutionary biology and genetics for many years. He explains that the axolotl’s ability to grow new limbs is not their only appealing characteristic as an animal model. Axolotls are tetrapods and share homologous structures with humans, such as feet and digits—a desirable trait for modeling the regeneration of appendages. Also, axolotls lay hundreds of exceptionally large eggs that are easy to manipulate and observe during experiments. Because of the work of Dr. Voss and others, the axolotl genome is well defined, enabling genome-wide studies of the events triggered by tissue damage.
Building an Axolotl Genome Assembly

It is vital to understand how the axolotl genome functions during regeneration. Stimulating or inhibiting genetic factors essential to tissue regrowth may lead to treatments in regenerative medicine. Recently, a research team led by Dr. Voss in collaboration with Dr. Jeremiah Smith, University of Kentucky, completed a chromosome-level assembly of the axolotl genome by aligning DNA scaffolds to a genetic linkage map. The map was created by crossing the axolotl to a closely related species, the eastern tiger salamander (Ambystoma tigrinum).2 The axolotl genome, at 32 billion base pairs, is the largest ever sequenced. It is approximately 10 times larger than the human genome.
Dr. Voss and his team continuously work to improve the genome assembly, which has considerable variation as a result of the inclusion of tiger salamander DNA. Dr. Voss explained that it is important to map more axolotls to identify which parts of the genetic code vary between individuals. The genes responsible for regeneration are likely to be highly conserved.
The axolotl genome assembly created by Dr. Voss and his colleagues is a boon to other researchers, enabling research in basic axolotl biology and providing a basis for gene expression studies and the development of molecular probes. Furthermore, similarities in the function of progenitor cells and cancer cells suggest that chemicals that arrest regeneration may be able to mitigate tumorigenesis as well.
Only a handful of laboratories currently have the expertise and equipment needed to use the genome assembly for genome-wide studies. During the next 5 years, Dr. Voss’s team will work to make the genome assembly data more accessible by improving annotation of genetic structures and identifying human homologous genes.
Gene Expression and Tissue Regeneration
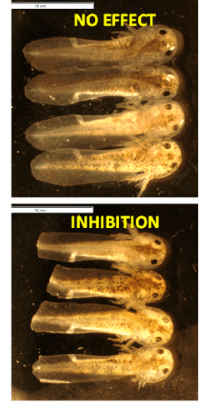
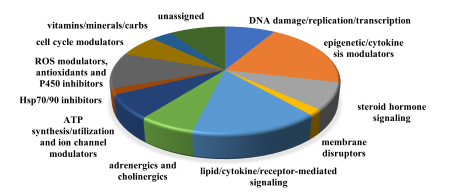
A concert of biochemical and genetic mechanisms must function perfectly for tissue regrowth to occur. Dr. Voss has been working in collaboration with Dr. Jon Thorson, Director of the Center for Pharmaceutical Research and Innovation at the University of Kentucky, to reveal details about the genetic and epigenetic factors driving regeneration. The team has developed an embryonic axolotl tail regeneration assay in which chemicals are applied to the growth medium of fertilized axolotl eggs. Two millimeters of the embryo’s tail are amputated at 20 days post‑fertilization, then researchers monitor tail regrowth to determine whether regeneration has been affected by the chemical. A non-treated embryo can complete tail tip regeneration in 7 days (Figure 3).3 Approximately 3,500 chemicals have been screened in the assay. Preliminary microarray analysis demonstrated altered transcription of a high number of diverse classes of genes (Figure 4). Effective compounds will be prioritized in future limb regeneration studies in adult axolotls.
If compounds affect regeneration, the team uses the mechanism of action of the chemical to trace back to the specific gene or genes involved. Because different genes are activated as the process of regeneration proceeds, different transcription factors may be affected at different time points. For example, embryonic exposure to the chemical C59 inhibited Wnt signal transduction pathways that are required for tail tip regeneration. Tissue analysis of RNA biomarkers at several time points after amputation showed that transcription was altered after C59 exposure, with a different array of genes affected at each time point.3
Dr. Voss and his team are using the CRISPR-Cas9 technique to demonstrate the phenotypic effect of removing certain genes. CRISPR-Cas9 is derived from a bacterial self-defense system. The endonuclease Cas9 makes site-specific double-stranded breaks in target DNA.4 The technique allows the axolotl genes involved in regeneration to be selectively removed, or “knocked out,” producing embryos
that lack the specific phenotypes associated with those genes. Early results of the tail regeneration assay using knockout embryos are promising. The Voss laboratory also is performing RNA-Seq analysis (a technique to identify and quantify sequences of RNA) on single cells and single nuclei to understand gene expression during regeneration in different cell types.
Looking to the Future
A great deal remains to be learned about axolotl biology and exactly what elements are involved in regeneration. In the future, phenocopying regenerative elements into a mammalian model might be possible, bringing researchers one step closer to applications in regenerative medicine. Videos and more information on AGSC may be found at https://ambystoma.uky.edu/genetic-stock-center/.
References
1 Crowner A, Khatri S, Blichmann D, et al. Rediscovering the axolotl as a model for thyroid hormone dependent development. Front Endocrinol (Lausanne). 2019;10:237. doi:10.3389/fendo.2019.00237.
2 Smith JJ, Timoshevskaya N, Timoshevskiy VA, et al. A chromosome-scale assembly of the axolotl genome. Genome Res. 2019; 29(2):317–324. doi:10.1101/gr.241901.118.
3 Ponomareva LV, Athippozhy AT, Thorson JS, et al. Using Ambystoma mexicanum (Mexican axolotl) embryos, chemical genetics, and microarray analysis to identify signaling pathways associated with tissue regeneration. Comp Biochem Physiol C Toxicol Pharmacol. 2015;178:128–135. doi:10.1016/j.cbpc.2015.06.004.
4 Jiang F, Doudna JA. CRISPR-Cas9 structures and mechanisms. Annu Rev Biophys. 2017;46:505–529. doi:10.1146/annurev-biophys-062215-010822.



