A 4D Atlas for Studying Cell Development in Worms
For more than 50 years, scientists have used the roundworm Caenorhabditis elegans (C. elegans)—a hermaphroditic nematode that is widely recognized as an important tool for genetics, cell biology, and neuroscience research. The entire adult nervous system and cell lineage of C. elegans was mapped in 19861 and since then, scientists have used this map of neuron connectivity to help elucidate diverse neurobiological mechanisms. Compared to the 100 billion neurons in the human brain, C. elegans has 302 neurons1 and has thousands of genes that are homologous to human genes. C. elegans has an intricate life cycle and a body that is transparent. Therefore, it is an ideal system to visualize spatial development throughout its life cycle and for modeling neurodegenerative diseases, such as Alzheimer’s.
It is important that scientists correctly interpret the complex behavior of cells during the various stages of development in a multidimensional plane. Given the fact that organisms exist in three-dimensional (3D) spaces naturally, microscopic techniques that capture development in the appropriate dimensional context are necessary. Imaging of embryos by conventional microscopy has been challenging due to the lack of spatial resolution. These challenges have hindered the ability to capture the precise position of each cell during development. Advances in microscopy2 have curbed some of these imaging problems, but have not resolved limitations that exist when combining data images from multiple embryos, navigating through hundreds of images over a span of hours, or tracking individual cells when the worm is undergoing morphological changes, which includes twisting of the body and muscle contracting. Supported by the National Institutes of Health, C. elegans researchers have designed technology that computationally tracks the position of cells and combines the data from multiple embryos to accurately model embryonic development in four dimensions (4D).3
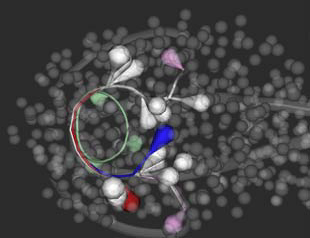
Leveraging this approach, a resource that is well positioned to assist scientists in this new era of C. elegans imaging is Worm Global Understanding in Dynamic Embryonic Systems (WormGUIDES).4 WormGUIDES is a project that encompasses advanced microscopy technology and various software that allows for automated and probabilistic cell tracking.5 Remarkably, the project presents a 4D atlas of the early developmental stages of C. elegans. The WormGUIDES atlas is an interactive representation of development and integrates dynamic imaging of cell positions and subcellular shape information through time. What’s interesting about the 4D atlas is that its technology can computationally manipulate the directionality of the worm embryo, “twisting” and “untwisting” it like spaghetti. The untwisting feature allows for scientists to better understand the formation of the C. elegans nervous system. The atlas has tools for data analysis and insight sharing. Users can annotate parts of the worm’s image to better understand biological functionality, which can be shared with other scientists. Image navigation is linked between 3D and lineage tree views. Developed in Java™ for Android™ and Objective-C for iOS™ (formerly iPhone OS), WormGUIDES is an open source5 for anyone with a smart device. This application is also available for desktop computers. The goal of WormGUIDES is to allow novel visual investigation of worm development by providing an interface for analyzing single cell level images of embryogenesis in 4D. This atlas has revolutionized the way that scientists examine developmental processes in C. elegans, resulting in newer hypotheses and greater insight into neuron development. In addition, this insight may aid in better understanding of the functionality of cells in the human nervous system.
References
1 White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 1986 November;314(1165):1-340.
2 Wu Y, Ghitani A, Christensen R, Santella A, Du Z, Rondeau G, Bao Z, Colón-Ramos D, Shroff H. Inverted selective plane illumination microscopy (iSPIM) enables coupled cell identity lineaging and neurodevelopmental imaging in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America 2011;108:17708–17713.
3 Christensen RP, Bokinsky A, Santella A, Wu Y, Marquina-Solis J, Guo M, Kovacevic I, Kumar A, Winter PW, Tashakkori N, McCreedy E, Liu H, McAuliffe M, Mohler W, Colón-Ramos DA, Boa Z, Shroff H. Untwisting the Caenorhabditis elegans embryo. eLife. 2015(4):e10070.
4 Santella A, Catena R, Kovacevic I, Shah P, Yu Z, Marquina-Solis J, Kumar A, Wu Y, Schaff J, Colón-Ramos, DA, Shroff H, Mohler WA, Bao Z. WormGUIDES: an interactive single cell developmental atlas and tool for collaborative multidimensional data exploration. BMC Bioinformatics. 2015 June 9(16):189.
5 http://www.wormguides.org/home



